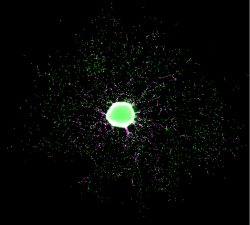
Neurodevelopmental toxicants are a serious risk for society, making regulatory testing for chemicals indispensable. Testing according to the current regulatory guidelines employing animal experiments is time- and cost intensive. For a more resource-effective way of developmental neurotoxicity (DNT) testing our laboratory established the human ‘Neurosphere Assay’, which is based on three dimensional (3D) neural progenitor cell (NPC) clusters. This system is able to mimic basic processes of brain development: Proliferation, differentiation, migration and apoptosis. Upon withdrawal of growth factors and in presence of an extracellular matrix (ECM), NPCs start to radially migrate out of the sphere-core and differentiate into astrocytes, neurons and oligodendrocytes. By immunostaining different cell types can be quantified and their morphology can be studied using high content image analyses (HCA).
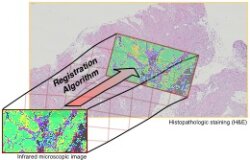
Measuring one and the same sample under two or more different types of microscopes is a common approach in medical diagnosis and other applications of vibrational microscpectroscopy. Often, the images from different microscopes need to be overlaid, so that each position in the sample has the same pixel coordinate in both images. This requires registration of the two images, which can be achieved using the Specreg software.
Click here to download the matlab source code.
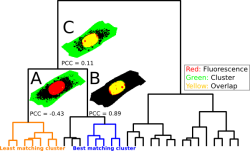
This colocalization scheme unveils statistically significant overlapping regions by identifying correlation between fluorescence color channels and clusters from unsupervised machine learning methods like hierarchical cluster analysis (HCA) performed on Raman or CARS spectral images. The scheme works as a pre-selection to gather appropriate spectra which can be used as training data to establish a supervised classifier (e.g. Random Forest) to automatically identify subcellular compartments.

Tracking cells in live cell imaging is an essential aspect of image analysis to quantify motility parameters in biological time-lapse microscopy. Cell tracking often gets complicated through background inhomogeneities and other side-effects of microscopic imaging. Ct3d implements a robust cell tracking algorithm based on a novel cosegmentation scheme that works reliable in 2D+t and 3D+t videos.

Many cell tracking algorithms have a tendency to oversegment or undersegment cells after a certain number of frames. Topaln corrects these missegmentation using a multiple matching approach that can be seen as a predecessor of the more systematic approach implemented in ct3d.
Description
topaln implements an integer linear programming approach for topologically aligning two segmented images to perform the linking step in cell tracking.
topaln is available under the GNU GPL, see Copyright.
Note that topaln requires CPLEX.
Download
To obtain topaln, please contact Axel Mosig by Email.

Fragrep implements a dynamic programming approach to efficient homology search for non-coding RNA genes. In its most recent version, this approach is combined with a structure-based search strategy.
BBQ is a command-line tool for discovering clusters of transcription factor binding sites that occur simultaneously in several genome sequences. Finding such clusters – which are sometimes also referred to as cis-regulatory modules – is done in a multiple-alignment-like fashion by solving a certain combinatorial and geometric optimization problem, the so-called best barbeque problem (explaining the name bbq). As opposed to classical, typically dynamic programming based, alignment procedures, the order of the binding sites' occurences can be arbitrarily shuffled, so that bbq is the result of developing completely new algorithms.